Writing eeglab Scripts
A script is a set of steps (functions) that are all called at once, and can be run on a batch of files. Writing scripts will save you hours, and will do repetitive tasks for you. Also, you can run multiple slow steps (such as ICA) back-to-back overnight.
For more info and another tutorial, here is the EEGLAB script writing tutorial.
Steps:
C R E A T E A N E W S C R I P T & A D D H E A D E R
C O P Y T H E F R A M E W O R K
D O T H E S T E P S W I T H T H E E E G L A B M E N U S
G E T T H E H I S T O R Y S C R I P T
R E P L A C E F I L E N A M E S W I T H V A R I A B L E S
Create a New Script & Add a Header
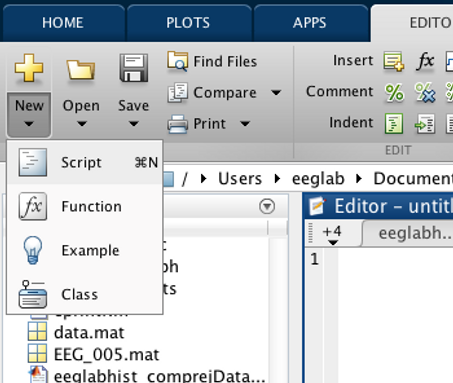
Creat a new Script. Click New --> Script OR type Command N.
Add an appropriate header.

Copy the Framework
Copy this framework and paste it into your own script, after your header. Copy and paste everything between the yellow lines. Use this framework anytime you want to be able to select one or more files to run a process on.
Example Code:
clear all
close all
%-----------------------These are the things you need to specify---------------------------------------------------------
% PUT THE THINGS THAT WILL BE THE SAME FOR EVERY FILE HERE. FOR EXAMPLE: A SAVE FOLDER OR THE LOCATION OF THE CHANNEL LOCATIONS FILE.
% EXAMPLE:
% exampleFolder = ('/Users/eeglab/Desktop/Post ICA Files 2/'); % Folder where finished files will be saved.
%
% CLpath = '/Users/eeglab/Documents/MATLAB/GSN256.sfp'; % The full path of the file containing the channel locations
% DEFINE YOUR VARIABLES HERE: ===========================================================================================
%========================================================================================================================
%------------------------------------------------------------------------------------------------------------------------
%% Ask for .raw file
% This function opens up a popup menu where you can click the files you
% want to run your function on. If you want to select multiple files, hold
% down command or shift while clicking.
% Get the files from a popup menu
% Use this function as [fileName, pathName] = uigetfile(filetype, title,
% defaultFolder, 'MultiSelect','on')
%
% filetype: * followed by the extention for the filetype you want, all in single quotes. Example: '*.raw' or '*.mat'
% title: the title of the popup window. You can put instructions to the user here, such as 'Select the .raw file(s).'
% defaultFolder: the path to the folder the menu opens up with. You can click to other folders from there. The path
% needs to be in single quotes. Example: '/Users/eeglab/Desktop/First Draft Data/After Comp Removal'
[fileName, pathName] = uigetfile('*.set', 'Select the file(s)', '/Users/eeglab/Desktop/First Draft Data/After Comp Removal', ...
'MultiSelect', 'on');
length = size(fileName);
if (iscellstr(fileName) == 0)
iMax = 1;
else
iMax = length(1,2);
end
for i = 1:iMax
if (iscellstr(fileName) == 0)
dataPath = strcat(pathName, fileName);
else
dataPath = strcat(pathName, char(fileName(1,i)));
end
% Parse the file path
[pathName,name,ext] = fileparts(dataPath); % pathstr = String containing the part of filename interpreted as a path name
% name = String containing the name of the file without any extension
% ext = String containing the file extension only, beginning with a period (.)
fileNameWithExt = strcat(name,ext); % Combine file's name and extention into one string called fileNameWithExt
% PUT YOUR CODE HERE: ===================================================================================================
%========================================================================================================================
end
This code is from framework.m. Your code goes between the sets of rows of equals signs.
Do the process with eeglab menus
(aka Graphic User Interface) If you're using EEGLAB functions, this step is a bit easier because EEGLAB will do some of the work for you.
Open (or close and reopen) eeglab. If you don't restart eeglab, you'll have extra functions show up in your history script.
Load or import a dataset.
To import a dataset, follow these instructions.
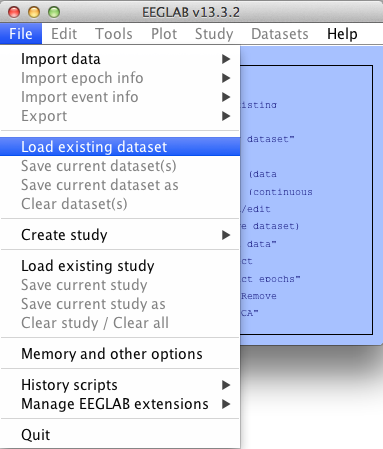
To load a dataset, in the EEGLAB window click File --> Load existing dataset
Do the process you want your function to do using the graphic interface (menus). For more info on these functions, see the other sections of this website, or the EEGLAB tutorials website. Carefully set the parameters in the menus. Most popup menus have the name of the function in the title bar (highlighted in gold):

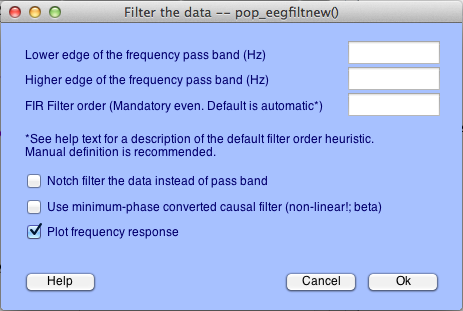

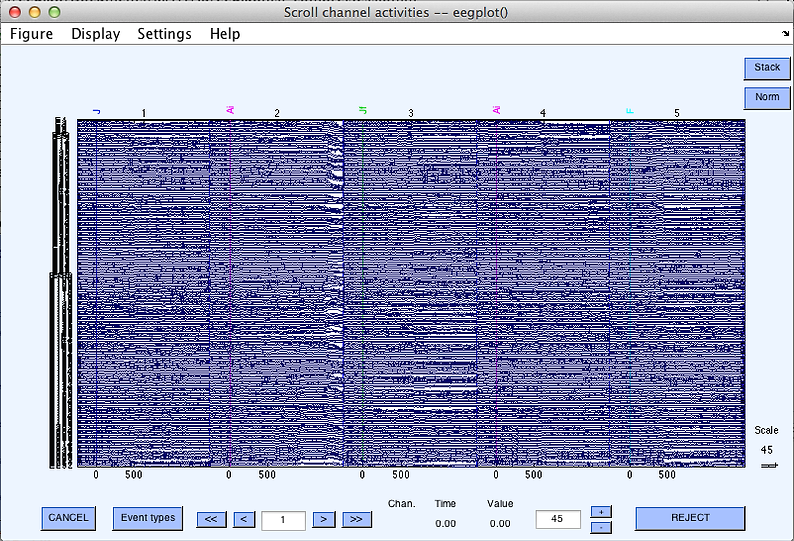
For popup menus like the first two above (pop_eegfiltnew and pop_topoplot), the parameters you type in the boxes (inside the blue boxes) will show up correctly formated as inputs to these functions in the history script. This'll make more sense when we get to the history script section.
Save the dataset. Click File --> Save current dataset as
History Script
Now save the session history script.
Click File -> History scripts -> Save session history script

Add a breif description to the file name. Note that the history script is saved to the MATLAB folder.

Now open the history file you just saved (fromthe MATLAB folder, which can be accessed in the left sidebar)
Copy and paste it into the framework in your script in the "PUT YOUR CODE HERE " section (almost at the bottom of the script).
Also, add this line of code right before the code you just pasted. This code opens EEGLAB.
[ALLEEG EEG CURRENTSET ALLCOM] = eeglab;

Adjust uigetfile function parameters
The uigetfile function makes a popup menu that allows you to select one or more files to run through your function.
Use this function as [fileName, pathName] = uigetfile(filetype, title, defaultFolder, 'MultiSelect','on')
filetype: * followed by the extention for the filetype you want, all in single quotes. Example: '*.raw' or '*.mat'
title: the title of the popup window. You can put instructions to the user here, such as 'Select the .raw file(s).'
defaultFolder: the path to the folder the menu opens up with. You can click to other folders from there. The path needs to be in single quotes. Example: '/Users/eeglab/Desktop/First Draft Data/After Comp Removal'
Examples:
[fileName, pathName] = uigetfile('*.set', 'Select the file(s)', '/Users/eeglab/Desktop/First Draft Data/After Comp Removal', 'MultiSelect', 'on');
[fileName, pathName] = uigetfile('*.raw', 'Select the .raw file(s)', '/Users/eeglab/Desktop/Pre ICA Files 2/', 'MultiSelect', 'on');

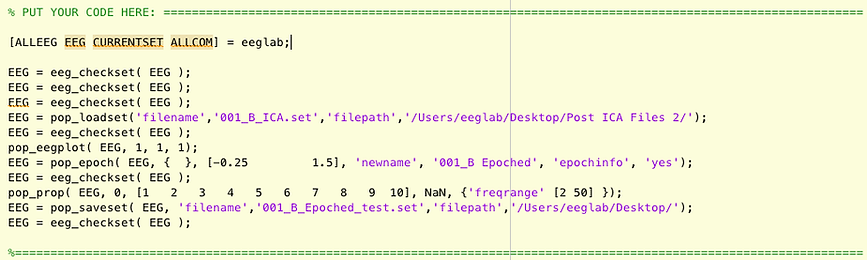
Now add comments that describe what the functions are accomplishing. The functions tend to have somewhat descriptive names, for example, pop_loadset loads the dataset, and pop_epoch epochs the data. Most of the EEGLAB functions have "eeg_" or "pop_" at the begining to designate that they are EEGLAB functions. If you're not sure what a function is doing, look it up in the documentation by clicking here. The eeg_checkset function makes sure the dataset is ok. Leave those where they are, but you don't need to specifically comment them.
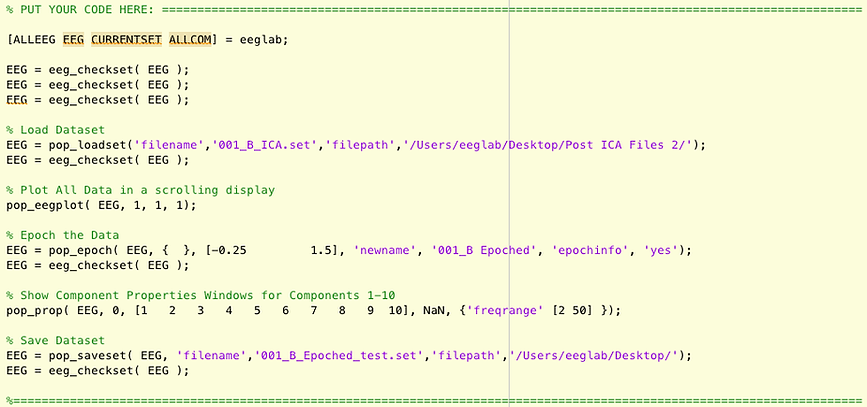
Replace filenames with Variables
You now need to replace some of the filenames with variables so that you are processing the correct file and naming it correctly.
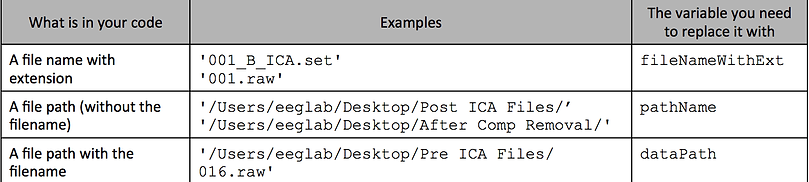
For file names & paths that refer to the file you selected/loaded/opened:
For file names & paths that refer to the name and place you saved the file
(after the processing)
Replace the path name with a varaible such as saveFolder, and define this variable in the "Define your variables here" section.
For the example in the video, I replace '/Users/eeglab/Desktop/' with saveFolder, and thenn I define saveFolder as '/Users/eeglab/Desktop/' in the DEFINE YOUR VARIABLES HERE section:




[
]
[
]
Replace the filename with strcat(name, '_yourTagHere.ext') where:
'_yourTagHere' is a brief description of what was done to the file. For example: '_ICA' or '_epoched'.
.ext is the extention that the file will be saved with. For example: .set or .mat



For dataset names
Anywhere the specific file name is, replace it with strcat(name, and the other parenthesis where appropriate.
For example: '001_B Epoched' --> strcat(name, ' Epoched').